loading things...
Projects
Three projects i have been meddling with.
Integrated Variant analysis of a pancreatic tumor in Galaxy and mapping against useful drugs for therapy in the Drug-gene database.
Jeremy Goecks, et al. Open pipelines for integrated tumor genome profiles reveal differences between pancreatic cancer tumors and cell lines, Cancer Medicine, Volume 4, Issue 3, 2015, Pages 392-403,https://doi.org/10.1002/cam4.360
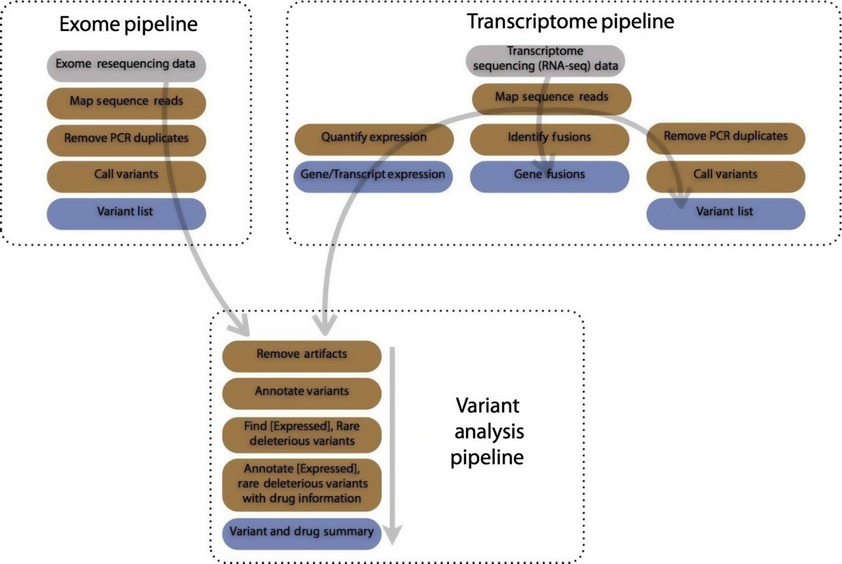
This project is built on three pipelines for personal oncology and integrated into Galaxy, a popular Web-based genomic workbench that supports pipelines. Collectively, these pipelines, an exome analysis pipe-line, a transcriptome (RNA-seq) analysis pipeline, and an integrated variant analysis pipeline, analyze a tumor sample to identify rare and deleterious mutations, druggable mutations, and drugs which may be effective for a tumor.
es.
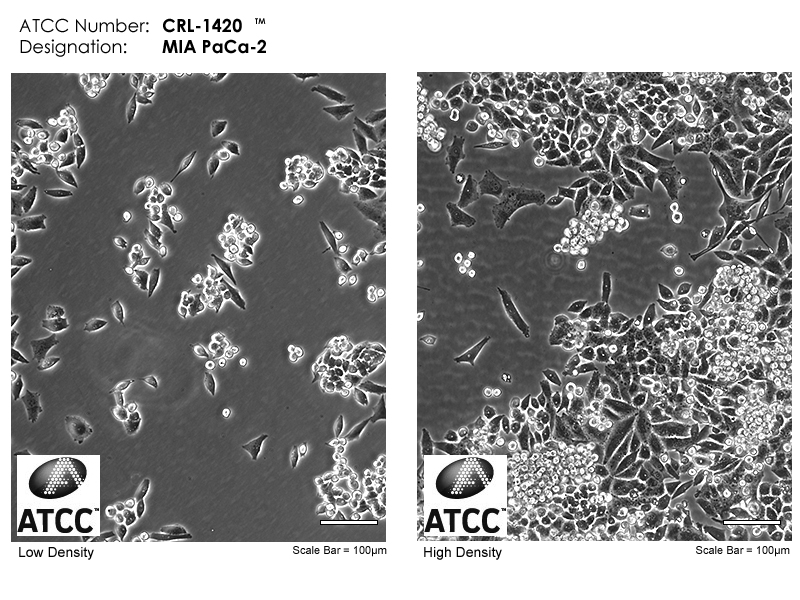
Tumor History:MIA PaCa-2 is a human pancreatic cancer cell line used extensively in pancreatic cancer research and therapy development. MIA PaCa-2 cells were derived from the carcinoma of a 65-year-old male.MIA PaCa-2 has served for decades as a model of pancreatic cancer, and studies of MIA PaCa-2 physiology have helped clarify the mechanisms of carcinogenesis in pancreatic cancer
Library Prep and Sequencing:
Total RNA from pancreatic cell lines and tumor tissue all had RIN>8.0 and were prepared using the Illumina TruSeq RNA kit (v1) according to manufacture's protocols. Final RNA‐Seq libraries were quantitated using qPCR and Agilent BioAnalyzer and sequenced using 100 bp paired‐end reads at 100,000 reads per sample with an Illumina HiSeq 2000 instrument.
#BWA, #SAMTOOLS, #PICARD, #VCFtools, #TOPHAT, #CUFFLINKS, #VARSCAN, #ANNOVAR
Below is a visual outline of the workflow
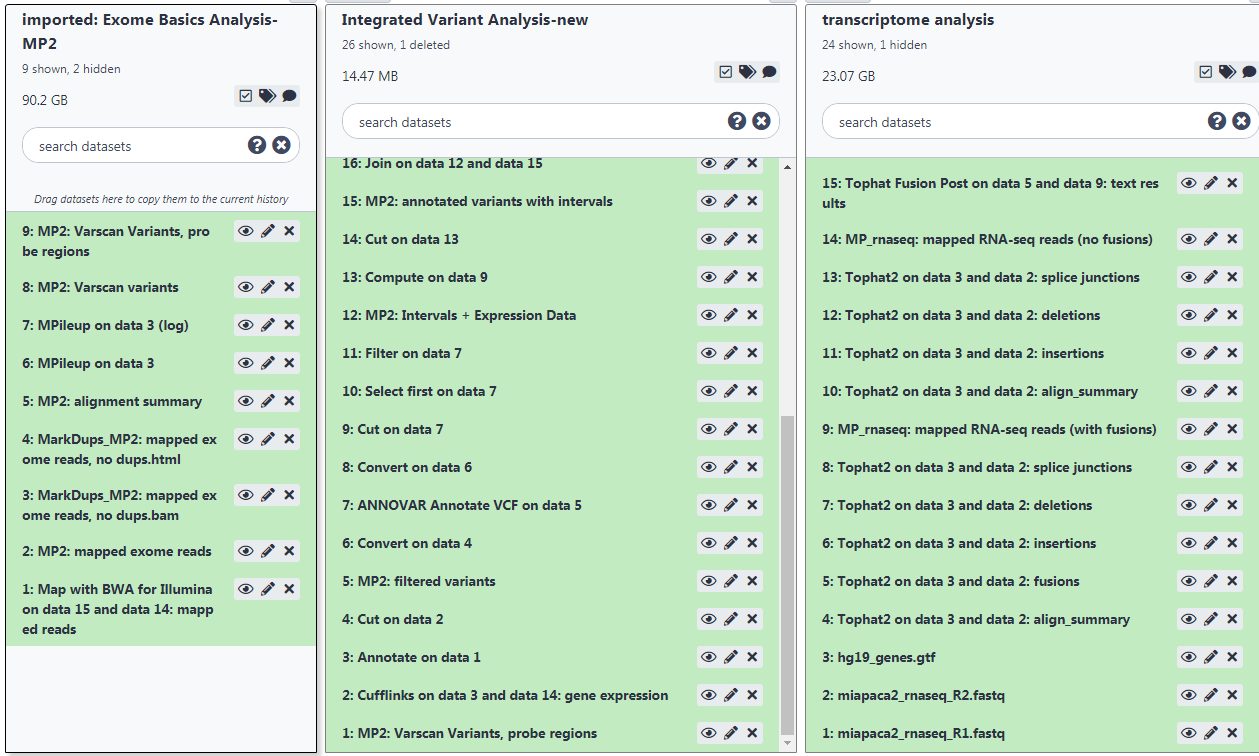
Here is a snap shot of the workflows running in Galaxy
There were a few technical problems running the experiment in Galaxy. All were resolved with the exception of the dgidb annotator tool that runs in step 21 of the integrated variant analysis pipeline.
The dgidb tool is responsible for annotating the tabular dataset obtained from ANNOVAR + expression annotated variants at step 19 with information from the drug-gene interaction database. I sent in a work ticket to the galaxy project tools/dev team to try and resolve the issue.
Unfortunately the problem was unable to be resolved at the time and so I basically self-fixed the problem by manually inputting my variant information into the drug-gene interaction database . Below is my tabular dataset obtained from running annovar.
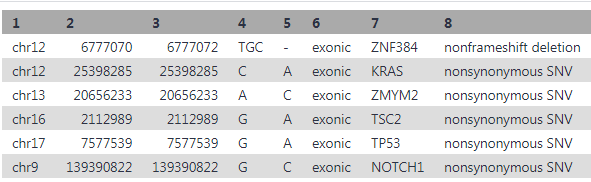
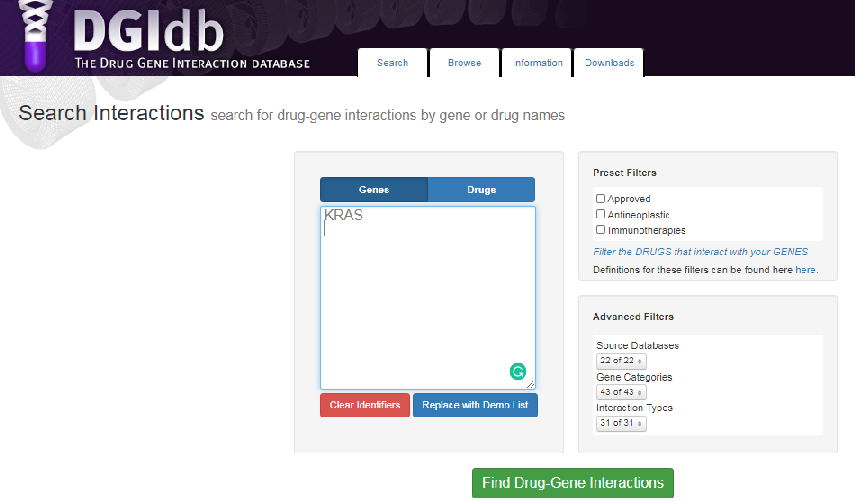
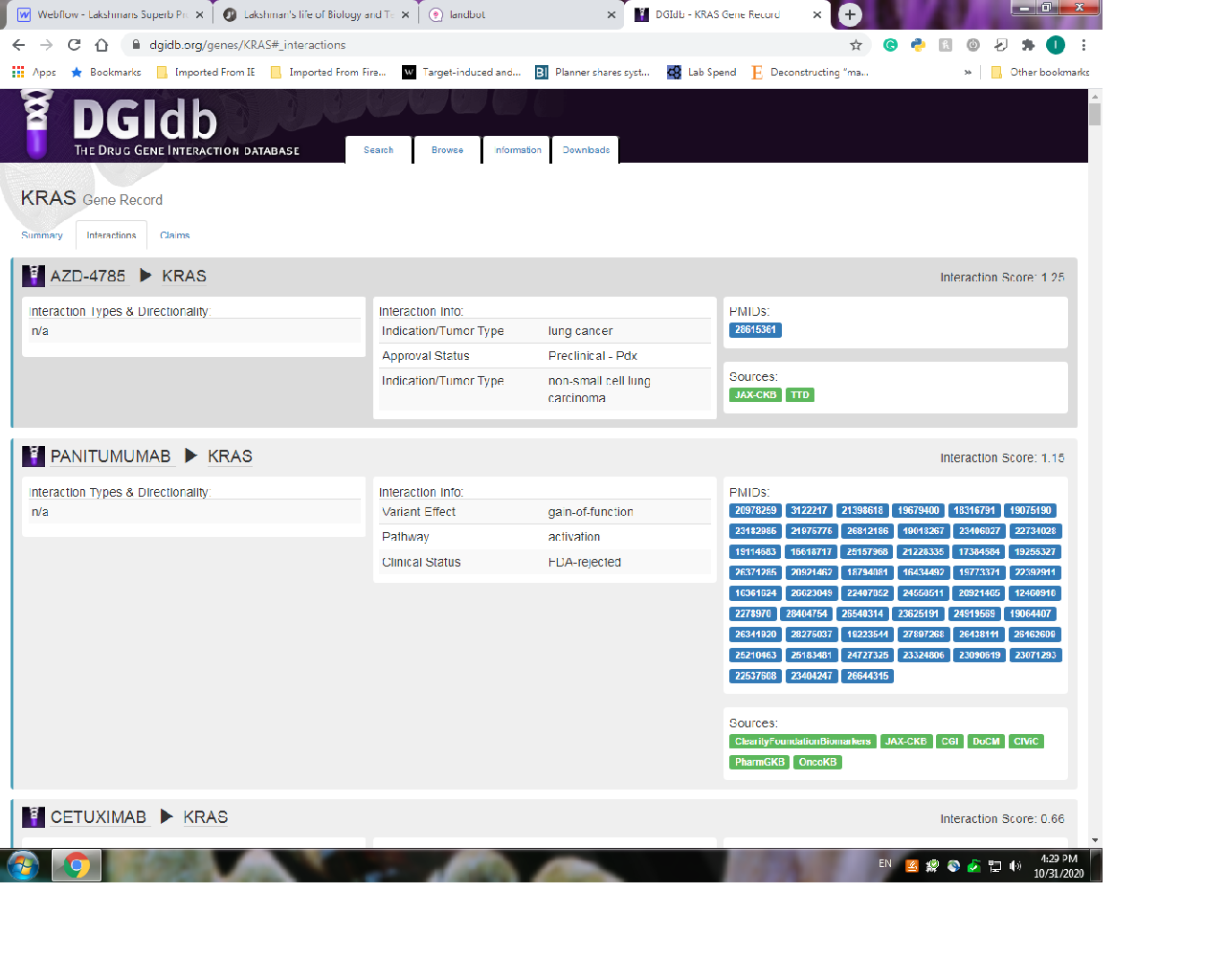
The table above shows chromosome number (Column 1), nucleotide location of mutation (Column's 2 & 3), base changes ( Column 4 & 5), DNA regions and name of genes containing mutations (col 6 &7) and the type of mutation (Col 8). With this information we can import the identified genes into the drug-gene interaction database @ https://www.dgidb.org/. Lets look at our first mutation in the table, specifically at the Kras gene. I will input the gene name into dgidb and look for potential drug-gene interactions and potential druggability. Below is a visual snap shot of the process.
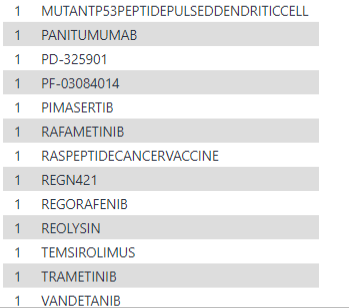
And so from the table, the gene names of the deleterious variants would serve as inputs. The outputs from the the DGI- database provides two critical pieces of information. First is a list of potential drug-gene interactions from our list of mutants. Second is a filtered down list showing actual potential "druggable" genes and the list of drugs that could have a therapeutic effect. Below is a list of the drugs!
Closing thoughts:
There is a lot more information i would like to share about this project, my personal thoughts on its usability, data and the step by step process for each of the pipelines. I will continue to add on to this page in the weeks to come! For access to the pipelines and data, Check my published galaxy page on the link below;
Galaxy-LinkBelow are two other projects that i initiated and are continuing to undergo development. The links below will take you to the github front pages, in which there are additional links that will direct you to the project files and the jupyter notebook.
An analysis in R & Python of the Plasmodium Falciparum parasite 3D7 Mitochondrion genome (Anopheles mosquito's are the natural hosts for the transmission of the parasite to humans causing malaria). Links to the project hosted on GitHub below.
GitHub
Neural network for breast cancer detection using Tensor flow in Google Colab. Links to the project hosted on GitHub below.
GitHub